Over 10 years experience of Traceability Solutions
By Pharmatrax Author
Category: Technoloy
 No Comments
No Comments
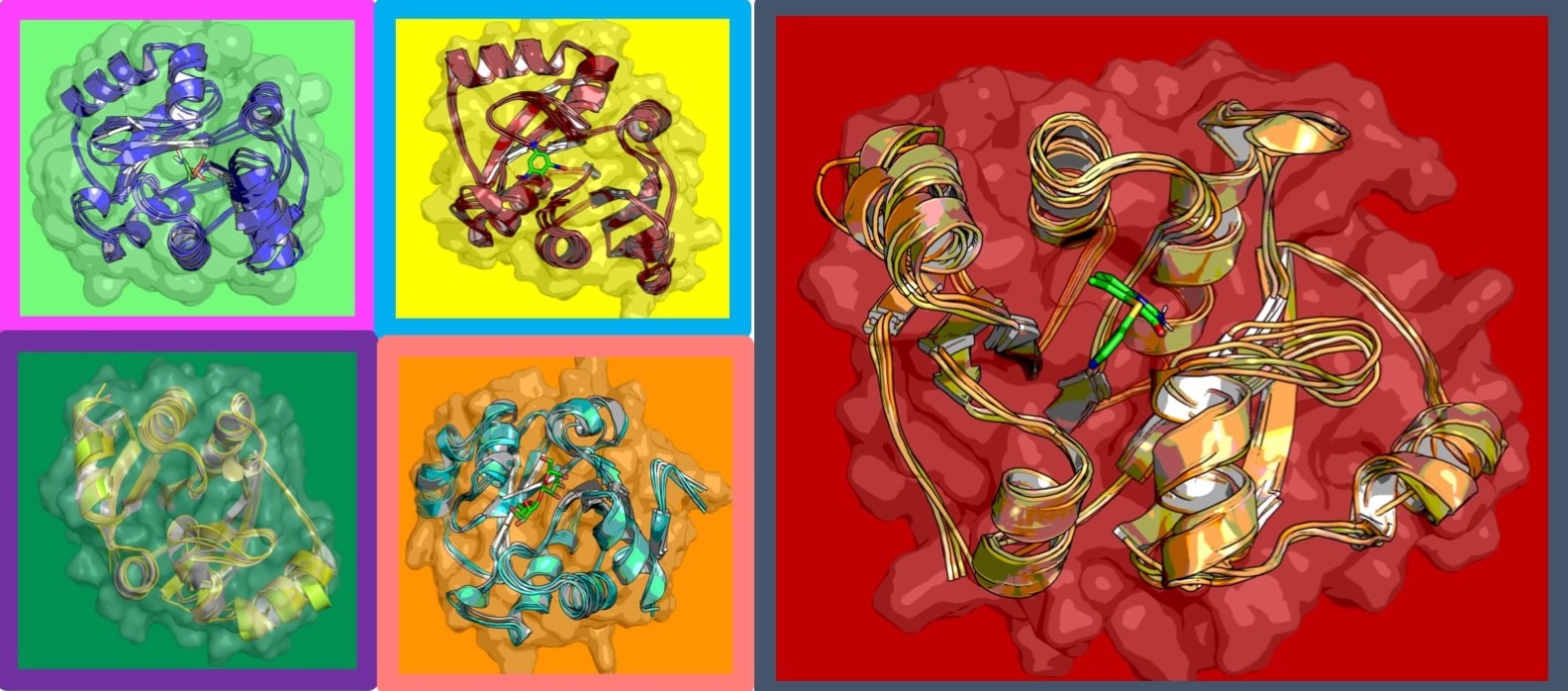
The world’s supercomputers are engaged in an urgent scavenger hunt, poring over as many molecules as possible in the hopes of finding one that bonds to COVID-19 effectively enough to be used as a drug. There’s a daunting backlog of molecules that need testing, however – numbering in the billions. Now, researchers at Argonne National Laboratory are leveraging supercomputing-powered AI to fast-track identification of the most promising molecules.
“We’re trying to build infrastructure to integrate AI and machine learning tools with physics-based tools,” explained Arvind Ramanathan, a computational biologist in the Data Science and Learning division at Argonne National Laboratory, in an interview with TACC’s Aaron Dubrow. “We bridge those two approaches to get a better bang for the buck.”
The research team used DeepDriveMD (short for Deep Learning-Driven Adaptive Molecular Simulations for Protein Folding), a tool originally developed for the Exascale Computing Project. DeepDriveMD was being adapted for cancer drug analysis when the pandemic hit, after which the researchers pivoted to COVID-19 analysis. DeepDriveMD starts with a simple protein molecule model and gradually complicates the model with new factors and more complex analyses, allowing the researchers to use deep learning to discover the aspects of protein that make them stronger candidates for COVID-19 binding.
“We built the toolkit to do the deep learning online, enabling it to sample as we go along,” Ramanathan said. “We first train it with some data, then allow it to infer on incoming simulation data very quickly. Then, based on the new snapshots it identifies, the approach automatically decides if the training needs to be revised.”
To train and run these heavy-duty models, the researchers turned to not one, not two, but four supercomputers: the 2.8 peak petaflop Comet system at the San Diego Supercomputer Center (SDSC); the 2.3 Linpack petaflop Longhorn system at the Texas Advanced Computing Center (TACC); the 23.5 Linpack petaflop Frontera system, also at TACC; and finally, the 148.6 Linpack petaflop Summit system at Oak Ridge National Laboratory (ORNL), which rated as the most powerful publicly ranked supercomputer in the world on the most recent Top500 list.
“TACC has been critical for our work, especially the Frontera machine,” Ramanathan said. “We’ve been going at it for a while, using Frontera’s CPUs to the maximum capacity to rapidly screen: taking virtual molecules and putting them next to a protein to see if it binds, and then infer from it whether other molecules will also do the same.” (Currently, the team is simulating 300,000 ligands per hour on Frontera.)
Using DeepDriveMD, the researchers drilled down from a billion molecules to a quarter billion to six million to a few thousand, eventually settling on 30 molecules with the greatest binding abilities. Those results are being shared with research collaborators and will soon be published in an open access report. Now, the researchers are moving on to analysis of the COVID-19 main protease and larger, more complicated proteins.
“In times of global need like this, it’s important not only that we bring all of our resources to bear, but that we do so in the most innovative ways possible,” said Dan Stanzione, TACC’s executive director. “We’ve pivoted many of our resources towards crucial research in the fight against COVID-19, but supporting the new AI methodologies in this project gives us the chance to use those resources even more effectively.”